Single-cell omics
single cell RNASeq data analysis with application to neurodegenerative diseases
Single-cell sequencing is a fast-growing technology in biology. It enables us to study the mechanism of biogical processes and diseases at single cell level. Single-cell RNA sequencing (scRNASeq) measures gene expression in each individual cell, allowing for identifying novel cell types; single-cell DNA sequencing can detect somatic mutations, which can be used to trace the evolution of cell lineages; single-cell ATAC-Seq can identify regulatory regions in each cell, in order to infer cell-type specific transcriptional networks. We are working on developing statistical tools to analyze single-cell sequencing data and integrate different omics to study the dynamics of cell states.
A main challenge in analyzing scRNA-seq data is to reduce technical variations yet retain cell heterogeneity. Due to low mRNAs content per cell and molecule losses during the experiment (called ‘‘dropout”), the gene expression matrix has a substantial amount of zero read counts. Some of the zeros as technical artifacts because of “dropout” and others are biological true zero as the gene is not expressed in the cell. The idea of imputation is to use the gene expression of other similar cells and co-expressed genes (e.g. in the same pathway) to identify whether the zero is true biological zero or “dropout”, and impute the true gene expression for the latter case. Existing imputation methods treat either each cell or each gene as independently and identically distributed, which oversimplifies the gene correlation and cell type structure. We propose a statistical model-based approach, called SIMPLEs (SInglecell RNA-seq iMPutation and celL clustErings), which iteratively identifies correlated gene modules and cell clusters and imputes dropouts customized for individual gene module and cell type. Simultaneously, it quantifies the uncertainty of imputation and cell clustering via multiple imputations. We model the gene expression by a mixture of zero-inflated censored multivariate Gaussian distribution, where the zero component is for “dropout” event. We then model the co-expression of gene modules by a factor model.
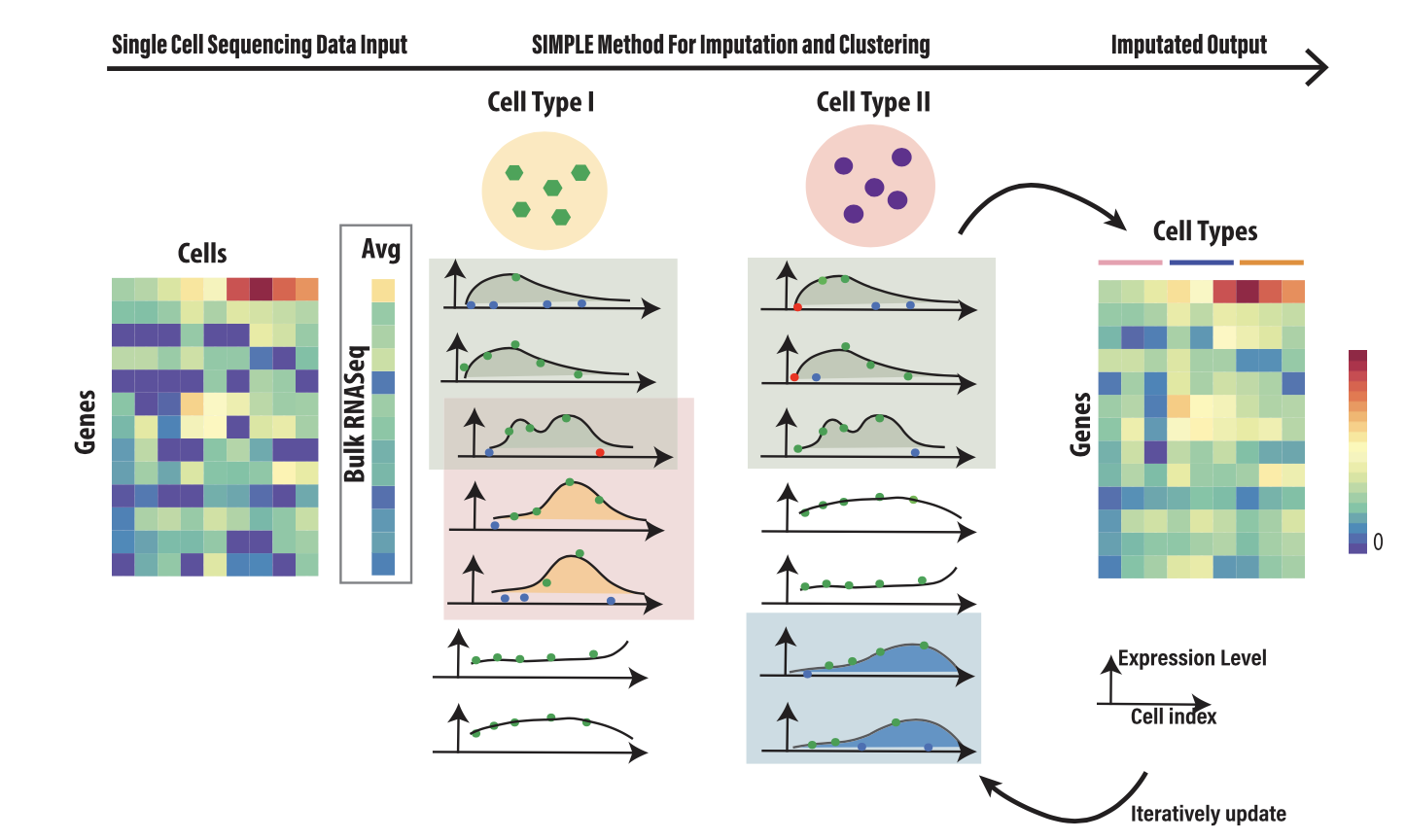
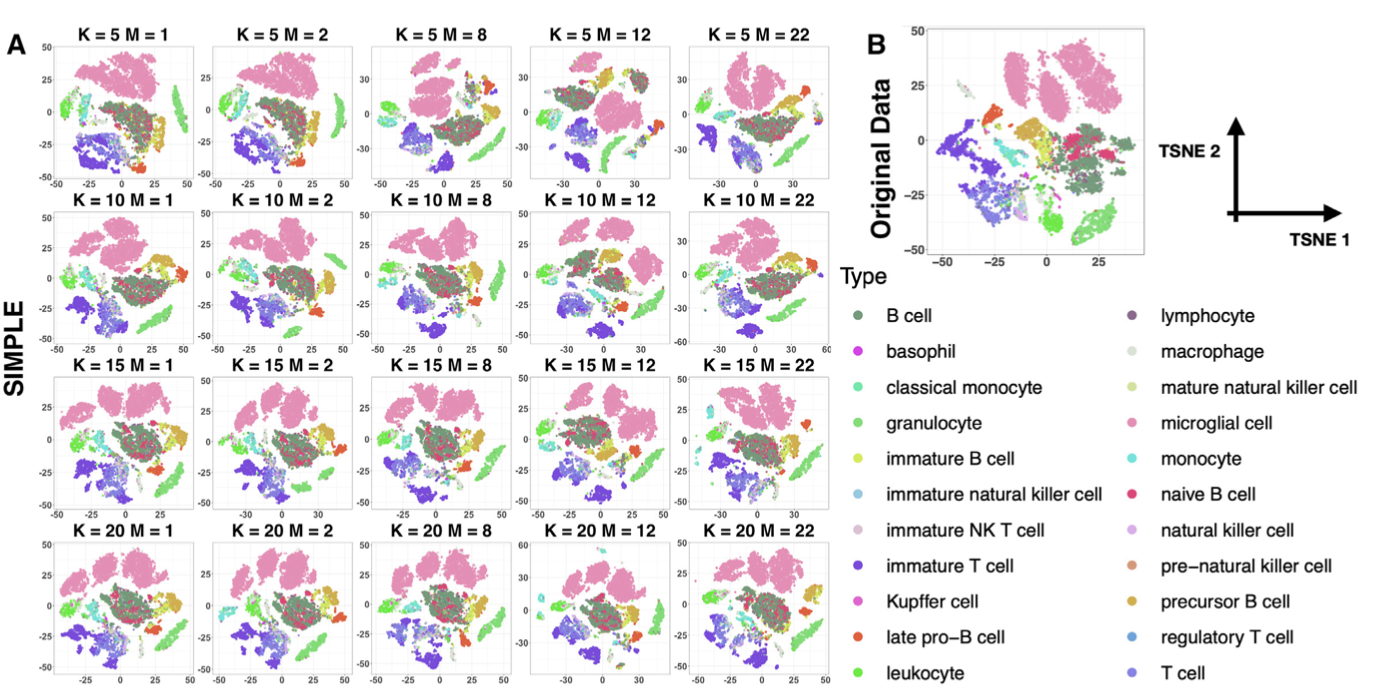
By applying SIMPLEs to several real datasets, we could identify cell clusters and discovered gene modules that can further classify subtypes of cells. Our imputations successfully recovered the expression trends of marker genes in stem cell differentiation and can discover putative pathways regulating biological processes.
Based on scRNASeq data, we study microglial-derived inflammation in neurodegenerative diseases and identify a subclass of microglia in mouse models of amyotrophic lateral sclerosis (ALS) which we term RIPK1-Regulated Inflammatory Microglia (RRIMs). RRIMs show significant up-regulation of classical proinflammatory pathways, and can be suppressed by inhibiting RIPK1. These findings help to elucidate a mechanism by which RIPK1 kinase inhibition has been shown to provide therapeutic benefit in mouse models of ALS and may provide an additional biomarker for analysis in ongoing phase 2 clinical trials of RIPK1 inhibitors in ALS. In the study of brain endothelial cells (EC) in Alzheimer’s Disease (AD), we find selective reduction of venous and capillary ECs in AD.

References
-
Zhirui Hu, Songpeng Zu, Jun S Liu. SIMPLEs: a single-cell RNA sequencing imputation strategy preserving gene modules and cell clusters variation. NAR Genomics and Bioinformatics, 2020, 2(4): lqaa077. Link
-
Lauren Mifflin, Zhirui Hu, Connor Dufort, Cynthia C Hession, Alec J Walker, Kongyan Niu, Hong Zhu, Nan Liu, Jun S Liu, Joshua Z Levin, Beth Stevens, Junying Yuan, and Chengyu Zou. A RIPK1-Regulated Inflammatory Microglial State in Amyotrophic Lateral Sclerosis. Proceedings of the National Academy of Sciences, 2021, 118 (13): e2025102118. Link
-
Chengyu Zou, Lauren Mifflin, Zhirui Hu, Tian Zhang, Bing Shan, Huibing Wang, Xin Xing, Hong Zhu, Xian Adiconis, Joshua Z. Levin, Fupeng Li, Chuan-Fa Liu, Jun S. Liu, Junying Yuan. Reduction of mNAT1/hNAT2 Contributes to Cerebral Endothelial Necroptosis and Aβ Accumulation in Alzheimer’s Disease. Cell Reports, 2020, 33(10): 108447. Link
-
SIMPLEs: single-cell RNA sequencing imputation and clustering. R package.