Bayesian phylogenetics
Bayesian methods for detecting convergence on phylogenetic tree

The conversation of DNA sequence indicates its important biological function. The gain and loss of conservation in some species can infer the function changes in evolutionary history. When the genomic region is conversed, its sequence has few substitutions to maintain the function; otherwise, its sequence accumulates substitutions when the function is lost. Phenotypic convergence, in which the same function or morphology evolves multiple times independently, often due to adaption to similar environmental changes, is generally believed to be a strong signature of natural selection. However, we generally do not have a robust understanding of the genomic changes underlying phenotypic convergence. A key question in evolutionary biology is that whether convergent phenotypes arise from repeated use of the same underlying genetic elements, or via independent genetic pathways. The increasing amount of whole genome sequencing and phenotypic data in various species facilitates the study of molecular basis underlying species evolution. However, it also requires sophisticated statistical modeling and efficient statistical computing methods.
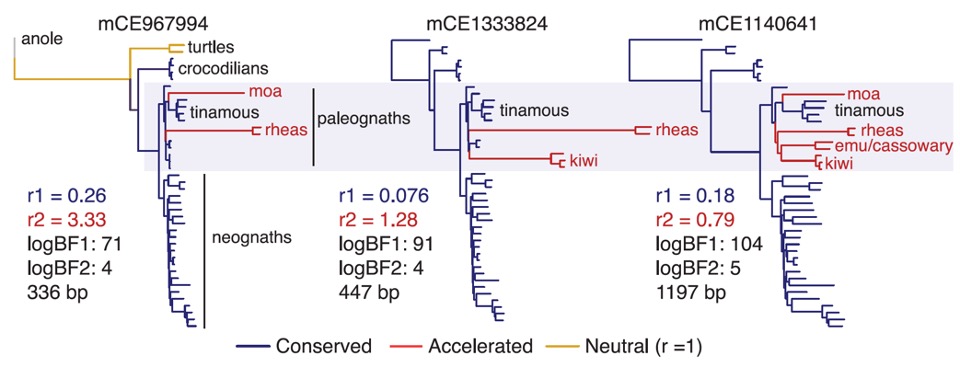
Convergence at the molecular level can arise via consistent shifts in substitution rate in genomic regions influencing particular phenotypic targets of natural selection. On a phylogenetic tree, the change of substitution rate on some branches can be used to detect the genomic regions that are associated with phenotypic convergence. However, existing methods do not allow easy detection of patterns of rate variation, which causes challenges for detecting convergent rate shifts or other complex evolutionary scenarios. Here we introduce PhyloAcc, a new Bayesian method to model substitution rate changes in conserved elements across a phylogeny. Compared with previous methods testing a predefined evolutionary history, PhyloAcc can distinguish different patterns of substitution rate changes. The method assumes several categories of substitution rate for each branch on the phylogenetic tree, estimates substitution rates per category, and detects changes of substitution rate as the posterior probability of a category switch. It uses Markov Chain Monte Carlo methods (MCMC) to estimate substitution rate shifts on the phylogeny, and computes Bayes factors for detecting genomic regions where rate shifts parallel with phenotype changes.
In two classic examples of convergent phenotypes, loss of flight in birds and the transition to marine life in mammals, our approach reveals numerous examples of conserved non-exonic elements with accelerations specific to the phenotypically convergent lineages. In the bird examples, we combined phylogenomic, developmental, and epigenomic analysis of 11 new genomes of paleognathous birds, including an extinct moa, to show that convergent evolution of regulatory regions, more so than protein-coding genes, is prevalent among developmental pathways associated with independent losses of flight. A Bayesian analysis of 284,001 conserved noncoding elements, 60,665 of which are corroborated as enhancers by open chromatin states during development, identified 2355 independent accelerations along lineages of flightless paleognaths, with functional consequences for driving gene expression in the developing forelimb. Our results suggest that the genomic landscape associated with morphological convergence in ratites has a substantial shared regulatory component. Our method is widely applicable to any set of conserved elements where multiple rate changes are expected on a phylogeny.
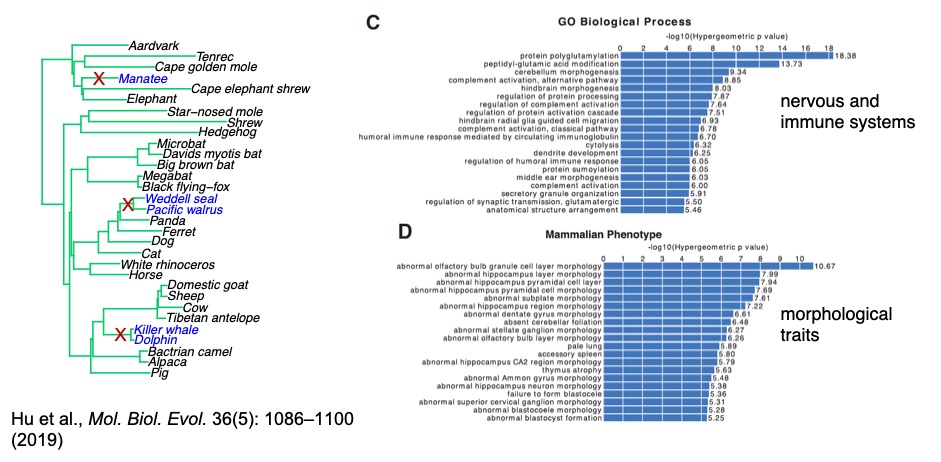
Another major issue not addressed by previous model is the possible heterogeneity in the gene trees across the genome. We later proposed PhyloAcc-GT, combining multispecies coalescent model with PhyloAcc framework to test the correlation between rate shifts and phenotypes per genomic region, accounting for phylogenetic uncertainty. PhyloAcc-GT uses MCMC to sample from the gene tree space, sums the likelihood over all probable gene trees given a species tree under the multispecies coalescent model, then compares the marginal likelihood under different hypotheses to identify genomic regions underlying phenotypic convergence. As the model becomes more complicated, it challenges current computational methods for statistical inference. We also improved several statistical computing methods, e.g., design more efficient proposal distribution for sampling gene trees and develop better methods for computing Bayes factor, which are critical problems in Bayesian statistics.
References
-
Zhirui Hu, Timothy B. Sackton, Scott V. Edwards, Jun S. Liu. Bayesian detection of convergent rate changes of conserved noncoding elements on phylogenetic trees. Molecular Biology and Evolution. 2019 36(5): 1086–1100. Link
-
Timothy B. Sackton, Phil Grayson, Alison Cloutier, Zhirui Hu, Jun S. Liu, Nicole E. Wheeler, Paul P. Gardner, Julia A. Clarke, Allan J. Baker, Michele Clamp, Scott V. Edwards. Convergent regulatory evolution and loss of flight in palaeognathous birds. Science 2019 364(6435):74-78. Link
-
PhyloAcc: a software to detect the changes of conservation of a genomic region. C/C++